Poster ID: FRI-208, EASL Congress 2024
ABSTRACT
Authors: David E Kleiner1, Andrew D. Clouston2, Zachary Goodman3, Cynthia D Guy4, Elizabeth M. Brunt5, Carolin Lackner6, Dina G. Tiniakos7, 8, Aileen Wee9, Matthew Yeh10, Wei Qiang Leow11, Elaine Chng12, Yayun Ren, George Boon Bee Goh13, Elizabeth E. Powell14, 15, Mary E. Rinella16, Arun J Sanyal17, Brent A. Neuschwander-Tetri18, Zobair Younossi19, Michael Charlton20, Vlad Ratziu21, Stephen A. Harrison22, 23, Dean Tai12, Quentin M. Anstee7, 24
1. Laboratory of Pathology; Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, United States
2. Molecular and Cellular Pathology, University of Queensland and Envoi Specialist Pathologists, Brisbane, Australia
3. Pathology Department, and Center for Liver Diseases, Inova Fairfax Hospital, Falls Church, Virginia, United States
4. Department of Pathology, Duke University Health System, Durham, NC, United States
5. Department of Pathology and Immunology, Washington University School of Medicine, Saint Louis, Missouri, United States
6. Institute of Pathology, Medical University of Graz, Graz, Australia
7. Translational and Clinical Research Institute, Faculty of Medical Sciences, Newcastle University, Newcastle upon Tyne, United Kingdom
8. Dept of Pathology, Aretaieion Hospital, National and Kapodistrian University of Athens, Athens, Greece
9. Department of Pathology, Yong Loo Lin School of Medicine, National University of Singapore, National University Hospital, Singapore, Singapore
10. Department of Pathology, University of Washington, Seattle, Washington, United States
11. Department of Anatomical Pathology, Singapore General Hospital, Singapore & Duke-NUS Medical School, Singapore, Singapore
12. HistoIndex Pte Ltd, Singapore, Singapore
13. Department of Gastroenterology and Hepatology, Singapore General Hospital, Singapore, Singapore
14. Centre for Liver Disease Research, Faculty of Medicine, University of Queensland, Translational Research Institute, Brisbane, Queensland, Australia
15. Department of Gastroenterology and Hepatology, Princess Alexandra Hospital, Brisbane, Queensland, Australia
16. Division of Gastroenterology and Transplant institute, Pritzker School of Medicine, University of Chicago, Chicago, Illinois, United States
17. Department of Internal Medicine, School of Medicine, Virginia Commonwealth University, Richmond, Virginia, United States
18. Division of Gastroenterology and Hepatology, Saint Louis University, Saint Louis, Missouri, United States
19. Betty and Guy Beatty Center for Integrated Research, Inova Health System, Falls Church, Virginia, United States
20. Center for Liver Diseases, and Transplantation Institute, University of Chicago, Chicago, Illinois, United States
21. Department of Hepatology, Sorbonne University and Pitié-Salpêtrière Hospital, Paris, France
22. Pinnacle Clinical Research, San Antonio, Texas, United States
23. Hepatology, Radcliffe Department of Medicine, University of Oxford, Oxford, United Kingdom
24. Newcastle NIHR Biomedical Research Centre, Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle upon Tyne, United Kingdom
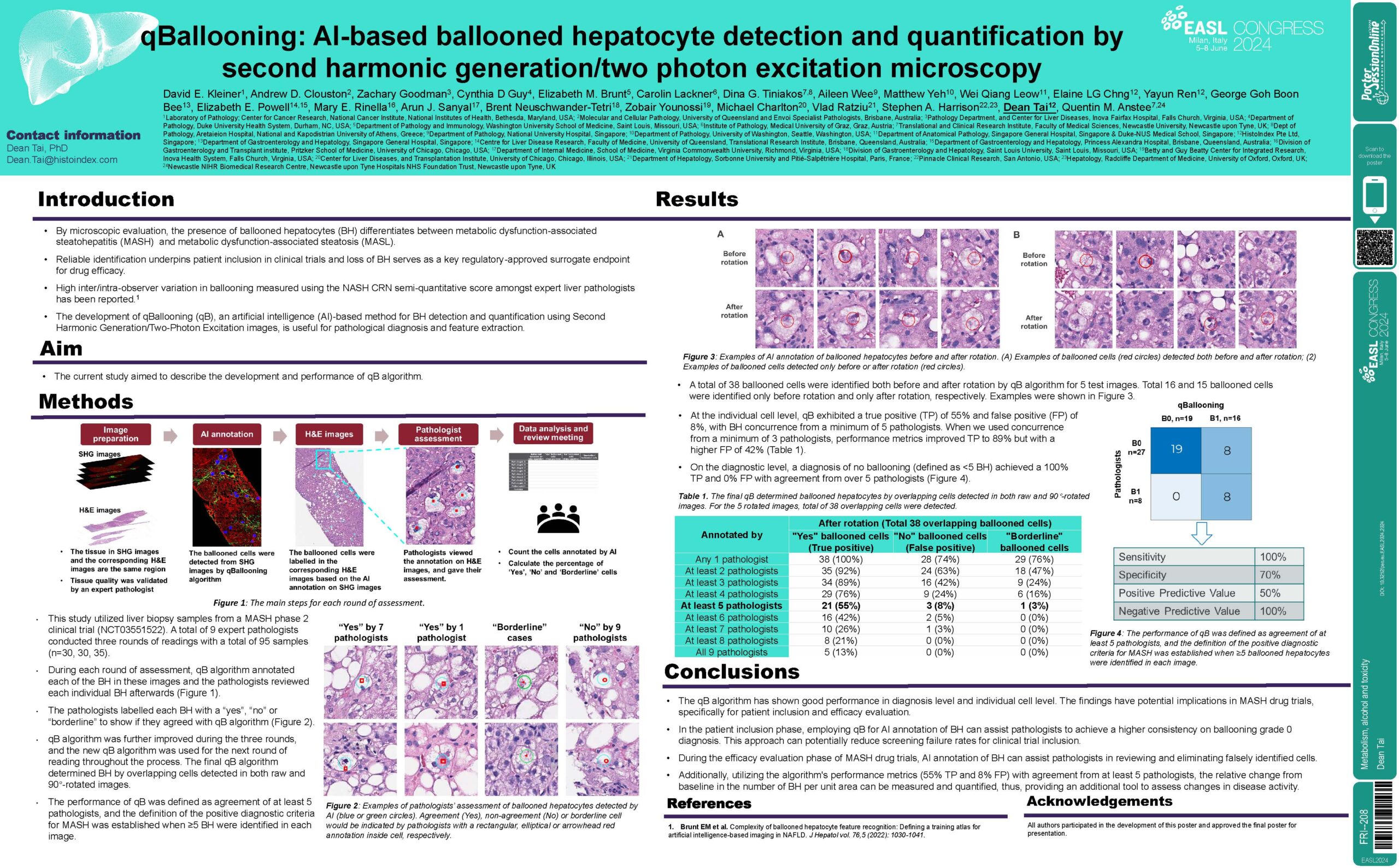
Background and Aims: Presence of ballooned hepatocytes (BH) differentiates between metabolic dysfunction-associated steatotic liver (MASL) and metabolic dysfunction-associated steatohepatitis (MASH). The development of qBallooning (qB), an artificial intelligence (AI)-based method for ballooned hepatocytes detection and quantification using Second Harmonic Generation and Two Photon Excitation images, is useful for pathological diagnosis and feature extraction. This study describes the development and performances of qB algorithm.
Method: A total of 9 expert pathologists conducted three rounds of readings with a total of 95 images (n=30, 30, 35). qB annotated each of the BH in these images and the pathologists reviewed each individual BH afterwards. The pathologists labelled each BH with a yes or no to show if they agreed with qB. qB was further improved during these three rounds, and the new qB was used for the next round of reading throughout the process. The final qB determined BH by overlapping cells detected in both raw and 90°-rotated images. The performance of qB was defined as agreement of at least 5 pathologists, and the definition of the positive diagnostic criteria for MASH was established when ≥5 BH were identified in each image.
Results: At the individual cell level, qB exhibited a true positive (TP) of 55% and false positive (FP) of 8%, with BH concurrence from a minimum of 5 pathologists. When we used concurrence from a minimum of 3 pathologists, performance metrics improved TP to 89% but with a higher FP of 42%. On the diagnostic level, a diagnosis of no ballooning (defined as <5 BH) achieved a 100% TP and 0% FP with agreement from over 5 pathologists.
Conclusion: The study’s findings have implications in MASH drug trial context, specifically in the patient inclusion and efficacy evaluation phases. In the patient inclusion phase, employing qB for AI annotation of BH can assist pathologists to achieve a higher consistency on ballooning grade 0 diagnosis. This approach can potentially reduce screening failure rates for clinical trial inclusion. During the efficacy evaluation phase of MASH drug trials, AI annotation on BH can assist pathologists in reviewing and eliminating falsely identified cells. Additionally, utilizing the algorithm’s performance metrics (55% TP and 8% FP) with agreement from at least 5 pathologists, the relative change from baseline in the number of BH per unit area can be measured and quantified thus providing an additional tool to assess changes in disease activity.